SM B: Effect Size Analysis
B.1 Meta-analysis
B.1.1 Effect Sizes \(Z_r\)
B.1.1.1 Effect of categorical review rating
The figures below (Figure B.1, Figure B.2) hows the fixed effect of categorical review rating on deviation from the meta-analytic mean. There is very little difference in deviation for analyses in any of the review categories. It is worth noting that each analysis features multiple times in these figures corresponding to the multiple reviewers that provided ratings.
Code
orchard_publishability <- function(dat){
rma_mod_rating <-
metafor::rma.mv(yi = Zr,
V = VZr,
data = dat,
control = list(maxiter = 1000),mods = ~ PublishableAsIs,
sparse = TRUE,
random = list(~1|response_id, ~1|ReviewerId))
orchaRd::orchard_plot(rma_mod_rating,
mod = "PublishableAsIs",
group = "id_col",
xlab = "Standardised Correlation Coefficient (Zr)",
cb = TRUE,angle = 45)
}
ManyEcoEvo_results$effects_analysis[[2]] %>%
filter(Zr > -4) %>%
unnest(review_data) %>%
select(Zr, VZr, id_col, PublishableAsIs, ReviewerId, response_id) %>%
mutate(PublishableAsIs = forcats::as_factor(PublishableAsIs) %>%
forcats::fct_relevel(c("deeply flawed and unpublishable",
"publishable with major revision",
"publishable with minor revision",
"publishable as is" ))) %>%
orchard_publishability() +
theme(text = element_text(size = 20),axis.text.y = element_text(size = 20)) +
scale_x_discrete(labels=c("Deeply Flawed\n & Unpublishable", "Publishable With\n Major Revision", "Publishable With\n Minor Revision", "Publishable\n As Is"))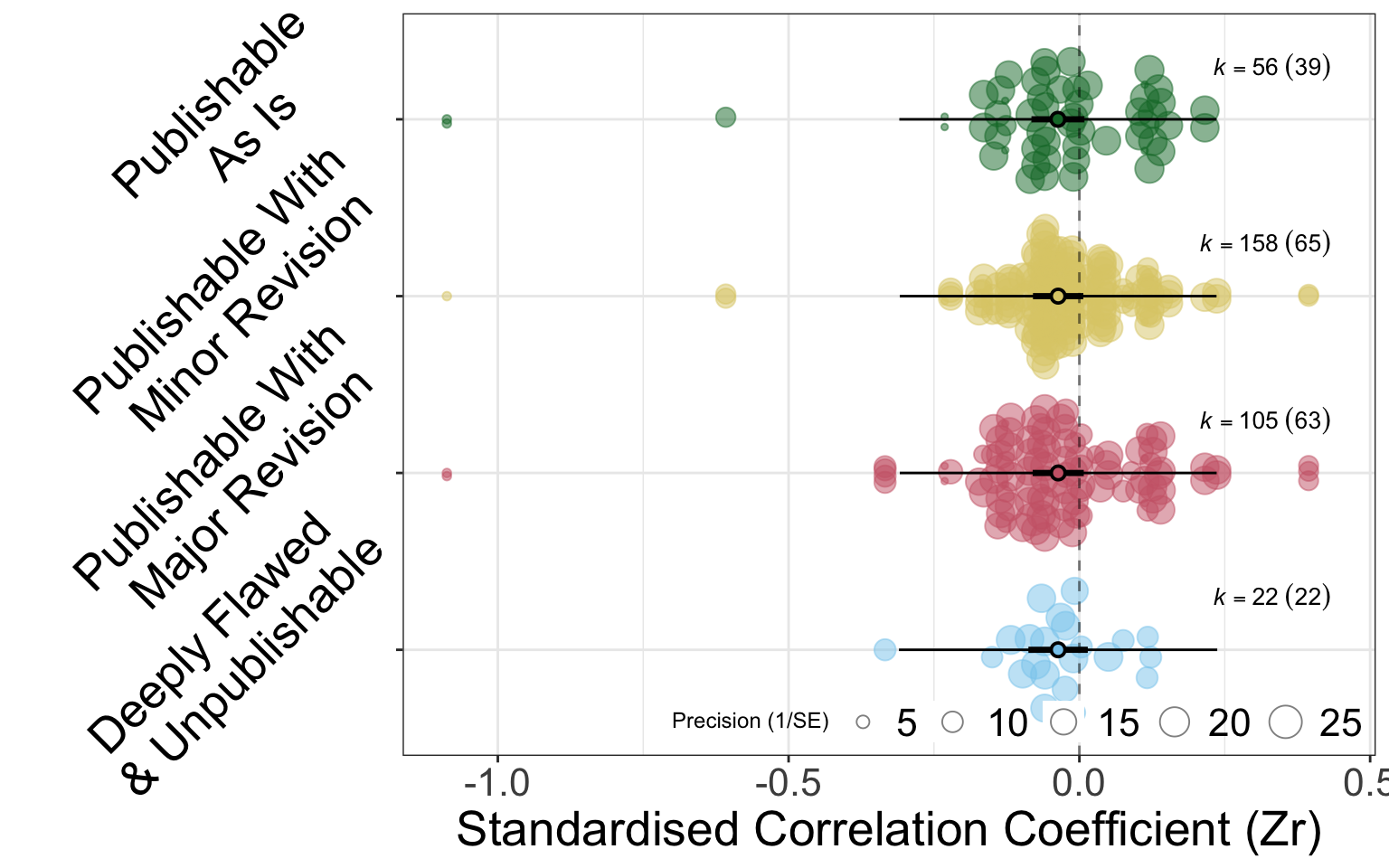
Code
ManyEcoEvo_results$effects_analysis[[1]] %>%
# filter(Zr > -4) %>%
unnest(review_data) %>%
select(Zr, VZr, id_col, PublishableAsIs, ReviewerId, response_id) %>%
mutate(PublishableAsIs = forcats::as_factor(PublishableAsIs) %>%
forcats::fct_relevel(c("deeply flawed and unpublishable",
"publishable with major revision",
"publishable with minor revision",
"publishable as is" ))) %>%
orchard_publishability() +
theme(text = element_text(size = 20),axis.text.y = element_text(size = 20)) +
scale_x_discrete(labels=c("Deeply Flawed\n & Unpublishable", "Publishable With\n Major Revision", "Publishable With\n Minor Revision", "Publishable\n As Is"))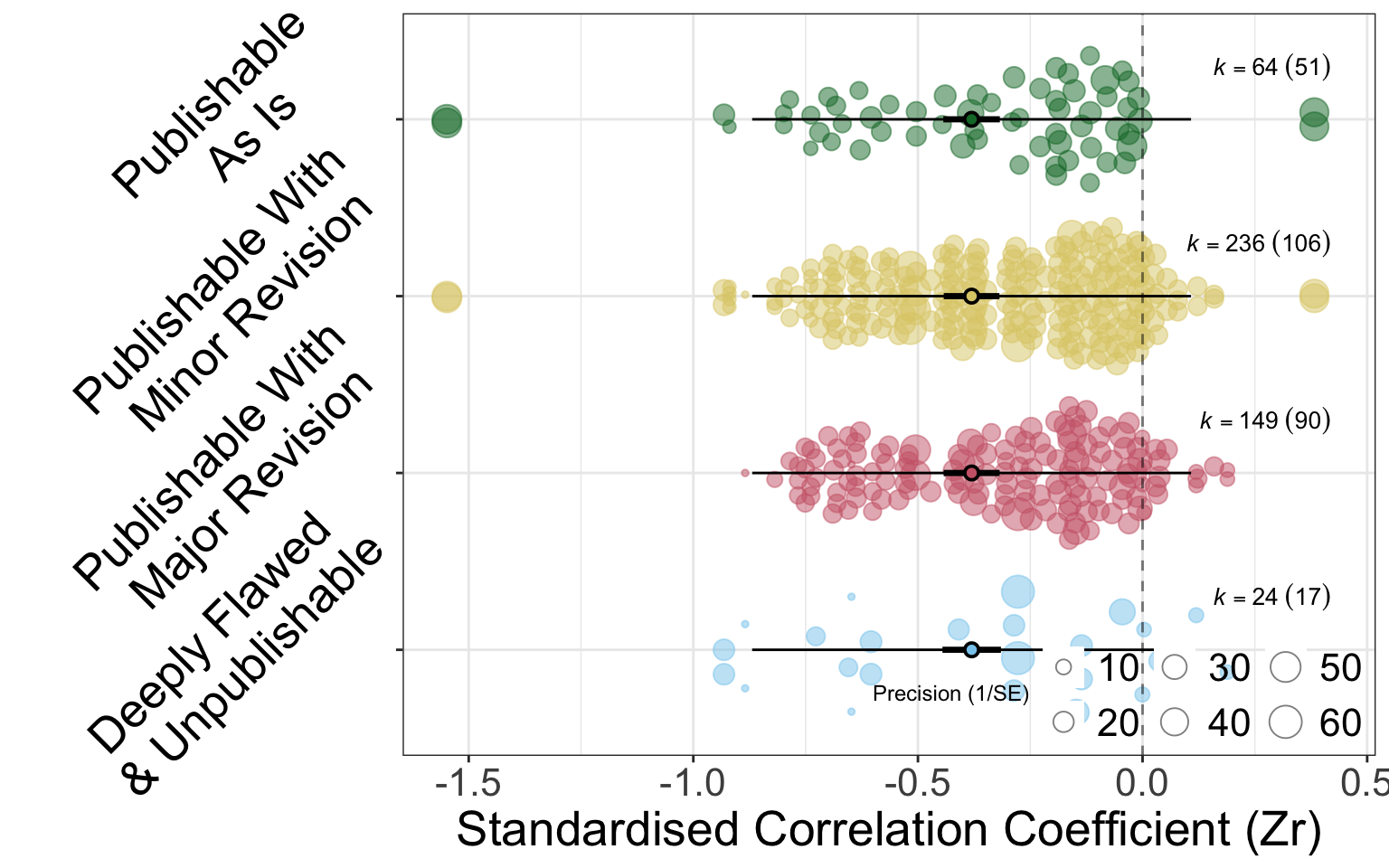
B.1.1.2 Post-hoc analysis: Exploring the effect of removing analyses with poor peer-review ratings on heterogeneity
The forest plots in Figure B.3 compare the distributions of \(Z_r\) effects from our full set of analyses with the distributions of \(Z_r\) effects from our post-hoc analyses, which removed either analyses that were reviewed at least once as being ‘unpublishable’, and analyses that were reviewed at least once as being ‘unpublishable’ or requiring ‘major revisions’. Removing these analyses from the blue tit data had little impact on the overall distribution of the results. When ‘unpublishable’ analyses of the Eucalyptus dataset were removed, the extreme outlier ‘Brooklyn-2-2-1’ was also removed, resulting in a substantial difference to the amount of observed deviation from the meta-analytic mean.
Code
# TeamIdentifier_lookup <- read_csv(here::here("data-raw/metadata_and_key_data/TeamIdentifierAnonymised.csv"))
plot_forest <- function(data, intercept = TRUE, MA_mean = TRUE){
if (MA_mean == FALSE){
data <- filter(data, Parameter != "overall")
}
p <- ggplot(data, aes(y = estimate,
x = term,
ymin = conf.low,
ymax = conf.high,
shape = point_shape,
colour = parameter_type)) +
geom_pointrange(fatten = 2) +
ggforestplot::theme_forest() +
theme(axis.line = element_line(linewidth = 0.10,
colour = "black"),
axis.line.y = element_blank(),
text = element_text(family = "Helvetica")#,
# axis.text.y = element_blank()
) +
guides(shape = "none", colour = "none") +
coord_flip() +
ylab(bquote(Standardised~Effect~Size~Z[r])) +
xlab(element_blank()) +
# scale_y_continuous(breaks = c(-4,-3,-2,-1,0,1),
# minor_breaks = seq(from = -4.5, to = 1.5, by = 0.5)) +
NatParksPalettes::scale_color_natparks_d("Glacier")
if(intercept == TRUE){
p <- p + geom_hline(yintercept = 0)
}
if(MA_mean == TRUE){
p <- p + geom_hline(aes(yintercept = meta_analytic_mean),
data = data,
colour = "#01353D",
linetype = "dashed")
}
return(p)
}
publishable_subsets_forest_data <-
ManyEcoEvo_viz %>%
filter(model_name == "MA_mod",
exclusion_set == "complete",
expertise_subset == "All") %>%
select(ends_with("set"), model, -expertise_subset) %>%
mutate(plot_data =
map(model,
.f =
~broom::tidy(.x,
conf.int = TRUE,
include_studies = TRUE) %>%
mutate(Parameter =
forcats::fct_reorder(term, estimate))),
meta_analytic_mean =
map_dbl(plot_data,
~ filter(.x,
Parameter == "overall") %>%
pull(estimate))) %>%
select(dataset,
publishable_subset,
plot_data,
meta_analytic_mean) %>%
unnest(cols = c("plot_data")) %>%
mutate(parameter_type =
case_when(
str_detect(Parameter, "overall") ~ "mean",
TRUE ~ "study")) %>%
group_by(dataset, publishable_subset) %>%
dplyr::mutate(point_shape =
ifelse(stringr::str_detect(term, "overall"),
"diamond",
"circle"))
# publishable_subsets_forest_data <-
# publishable_subsets_forest_data %>%
# rename(id_col = term) %>%
# group_by(type) %>%
# group_split() %>%
# set_names(., publishable_subsets_forest_data$type %>% unique) %>%
# # map_if(.x = ., names(.) == "study",
# # .f = ~ anonymise_teams(.x, TeamIdentifier_lookup)) %>%
# bind_rows() %>%
# rename(term = id_col)
library(tidytext)
tidy_overall_labeller <- . %>%
str_split("_") %>%
flatten_chr() %>%
pluck(1)
tidy_forest_labels <- Vectorize(tidy_overall_labeller)
publishable_subsets_forest_data %>%
group_by(dataset, publishable_subset) %>%
mutate(term = case_when(term == "overall" ~
paste(term,
dataset,
publishable_subset,
sep = "_"),
TRUE ~ term),
dataset = case_when(dataset == "blue tit" ~ "Blue tit",
dataset == "eucalyptus" ~ "Eucalyptus",
TRUE ~ NA)) %>%
arrange(across(.cols = c(type, estimate)),
.by_group = TRUE) %>%
rowid_to_column() %>%
mutate(term = reorder(term, rowid),
publishable_subset =
case_when(publishable_subset == "All" ~
"All analyses",
publishable_subset == "data_flawed" ~
"'Unpublishable'\nremoved",
publishable_subset == "data_flawed_major" ~
"'Unpublishable' &\n'Major Revisions'\nremoved",
TRUE ~ "")) %>%
plot_forest() +
scale_x_reordered(labels = tidy_forest_labels) +
ggh4x::facet_nested(dataset ~ publishable_subset,
independent = "y",
scales = "free")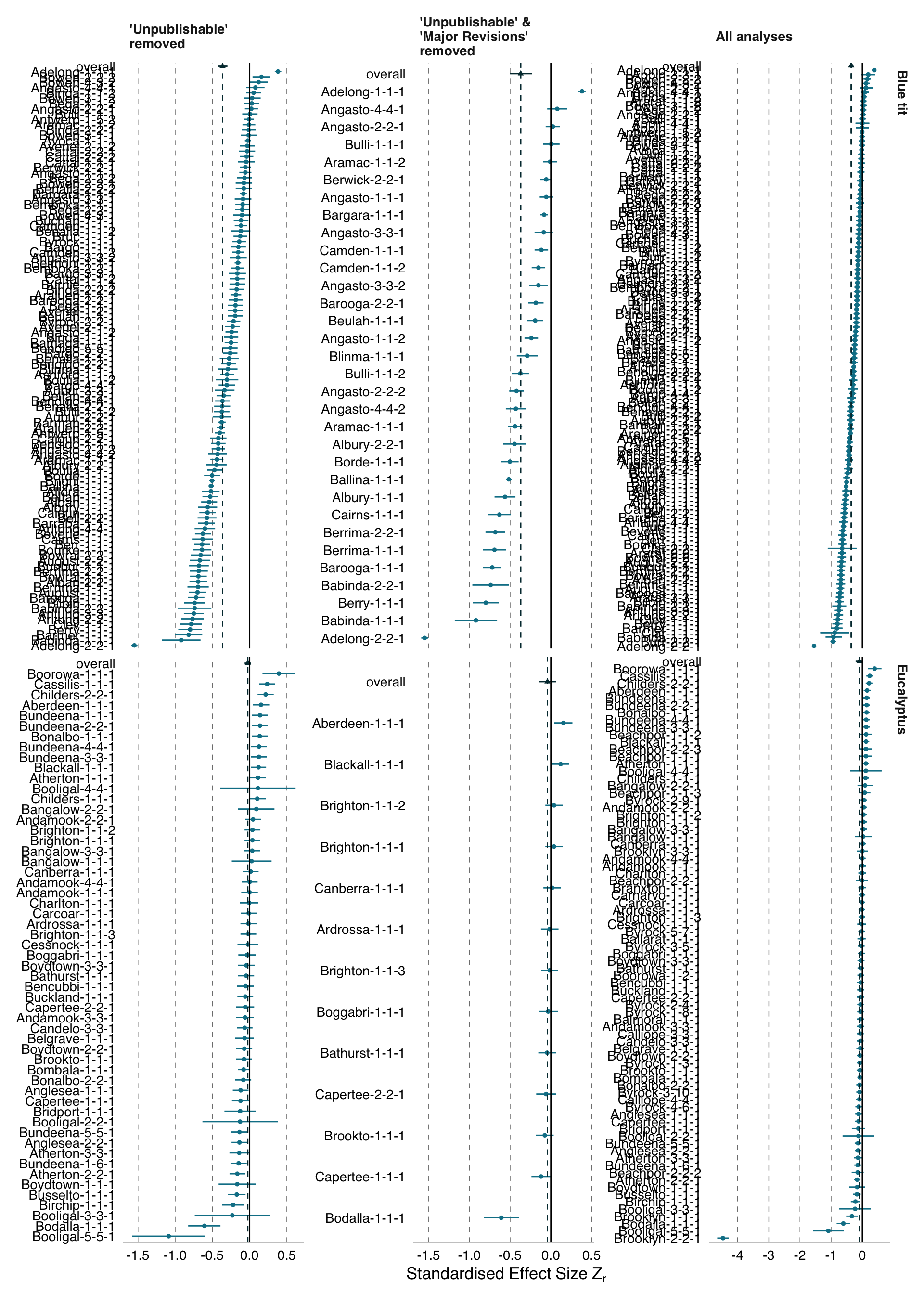
B.1.1.3 Post-hoc analysis: Exploring the effect of excluding estimates in which we had reduced confidence
For each dataset (blue tit, Eucalyptus), we created a second, more conservative version, that excluded effects based on estimates of \(\mathit{df}\) that we considered less reliable (Table B.1). We compared the outcomes of analyses of the primary dataset (constituted according to our registered plan) with the outcomes of analyses of the more conservative version of the dataset. We also compared results from analyses of both of these versions of the dataset to versions with our post-hoc removal of outliers described in the main text. Our more conservative exclusions (based on unreliable estimates of \(\mathit{df}\) ) had minimal impact on the meta-analytic mean for both blue tit and Eucalyptus analyses, regardless of whether outliers were excluded (Table B.1).
Code
ManyEcoEvo_viz %>%
dplyr::filter(estimate_type == "Zr",
model_name == "MA_mod",
collinearity_subset != "collinearity_removed",
publishable_subset == "All",
expertise_subset == "All") %>%
select(dataset, exclusion_set, tidy_mod_summary) %>%
unnest(tidy_mod_summary) %>%
filter(type == "summary") %>%
select(-term, -type) %>%
mutate(exclusion_set =
case_when(exclusion_set == "complete" ~
"Primary dataset",
exclusion_set == "complete-rm_outliers" ~
"Primary dataset, outliers removed",
exclusion_set == "partial" ~
"Conservative exclusions",
TRUE ~ "Conservative exclusions, outliers removed")) %>%
group_by(exclusion_set) %>%
gt::gt() %>%
gt::opt_stylize(style = 6, color = "gray") %>%
gt::fmt(columns = "p.value",
fns = function(x) gtsummary::style_pvalue(x, prepend_p = FALSE)) %>%
gt::fmt_number(columns = c(-p.value, -dataset)) %>%
gt::cols_label(estimate = gt::md("$$\\hat\\mu$$"),
std.error = gt::md("$$\text{SE}[\\hat\\mu]$$"),
conf.low = gt::md("95\\%CI")) %>%
gt::cols_merge(columns = starts_with("conf"),
pattern = "[{1},{2}]") %>%
gt::cols_move(columns = conf.low, after = std.error) %>%
gt::tab_style(
style = list(gt::cell_text(transform = "capitalize"),
gt::cell_text(style = "italic")),
locations = gt::cells_body(columns = "dataset", rows = dataset == "eucalyptus")
) | dataset | \[\hat\mu\] |
\[ ext{SE}[\hat\mu]\] |
95%CI |
statistic | p.value |
|---|---|---|---|---|---|
| Primary dataset | |||||
| blue tit | −0.35 | 0.03 | [−0.41,−0.29] | −11.02 | <0.001 |
| eucalyptus | −0.09 | 0.06 | [−0.22,0.03] | −1.47 | 0.14 |
| Conservative exclusions | |||||
| blue tit | −0.36 | 0.03 | [−0.42,−0.29] | −10.77 | <0.001 |
| eucalyptus | −0.11 | 0.07 | [−0.24,0.03] | −1.55 | 0.12 |
| Primary dataset, outliers removed | |||||
| blue tit | −0.36 | 0.03 | [−0.42,−0.30] | −11.48 | <0.001 |
| eucalyptus | −0.03 | 0.01 | [−0.06,0.00] | −2.23 | 0.026 |
| Conservative exclusions, outliers removed | |||||
| blue tit | −0.36 | 0.03 | [−0.43,−0.30] | −11.38 | <0.001 |
| eucalyptus | −0.04 | 0.02 | [−0.07,−0.01] | −2.52 | 0.012 |
Code
plot_forest <- function(data, intercept = TRUE, MA_mean = TRUE ){
if (MA_mean == FALSE) {
data <- filter(data, term != "Overall")
}
p <- ggplot(data, aes(y = term,
x = estimate,
ymin = conf.low,
ymax = conf.high,
# shape = point_shape,
colour = parameter_type)) +
geom_pointrange() +
ggforestplot::theme_forest() +
theme(axis.line = element_line(size = 0.10, colour = "black"),
axis.line.y = element_blank(),
text = element_text(family = "Helvetica"),
axis.text.y = element_blank()) +
guides(shape = "none", colour = "none") +
coord_flip() +
labs(y = "Standardised Effect Size, Zr",
x = element_blank()) +
scale_x_continuous(breaks = c(-4,-3,-2,-1,0,1),
minor_breaks = seq(from = -4.5, to = 1.5, by = 0.5)) +
NatParksPalettes::scale_color_natparks_d("Glacier")
if (intercept == TRUE) {
p <- p + geom_hline(yintercept = 0)
}
if (MA_mean == TRUE) {
p <- p + geom_hline(aes(yintercept = meta_analytic_mean),
data = data,
colour = "#01353D",
linetype = "dashed")
}
return(p)
}Code
complete_euc_data <-
ManyEcoEvo_viz %>%
filter(exclusion_set == "complete",
estimate_type == "Zr",
model_name == "MA_mod",
dataset == "eucalyptus",
publishable_subset == "All",
expertise_subset == "All") %>%
select(tidy_mod_summary) %>%
mutate(plot_data = map(tidy_mod_summary,
.f = ~ dplyr::mutate(.x,
point_shape =
ifelse(stringr::str_detect(term, "overall"),
"diamond",
"circle"),
Parameter =
forcats::fct_reorder(term,
estimate) %>%
forcats::fct_reorder(.,
point_shape,
.desc = TRUE))
),
meta_analytic_mean = map_dbl(plot_data,
~ filter(.x, Parameter == "overall") %>%
pull(estimate))) %>%
select(plot_data, meta_analytic_mean) %>%
unnest(cols = c("plot_data")) %>%
mutate(parameter_type = case_when(str_detect(Parameter, "overall") ~ "mean",
TRUE ~ "study"))
# ManyEcoEvo_viz %>%
# filter(exclusion_set == "complete",
# estimate_type == "Zr",
# model_name == "MA_mod",
# dataset == "eucalyptus",
# publishable_subset == "All",
# expertise_subset == "All") %>%
# )
min_outlier_euc <- complete_euc_data %>%
filter(type == "study") %>%
slice_min(estimate, n = 3) %>%
pull(term)
sample_size_euc_Zr <- ManyEcoEvo_results %>%
filter(exclusion_set == "complete", dataset == "eucalyptus") %>%
pluck("data", 1) %>%
select(id_col, sample_size) %>%
rename(term = id_col) %>%
mutate(sample_size = as.numeric(sample_size))
mean_n_euc_Zr <- sample_size_euc_Zr %>%
drop_na(sample_size) %>%
pull(sample_size) %>%
mean() %>%
round(2)
N_outliers_Zr_euc <- sample_size_euc_Zr %>%
filter(term %in% min_outlier_euc) %>%
arrange(desc(sample_size))B.1.1.4 Post-hoc analysis: Exploring the effect of including only analyses conducted by analysis teams with at least one member self-rated as “highly proficient” or “expert” in conducting statitistical analyses in their research area
The anonymous Team Identifiers in the reduced subset of “expert” or “highly proficient” analysts are exported internally in the ManyEcoEvo package as ManyEcoEvo:::expert_subset. Analyses from the following teams are retained in the reduced subset: Bell, Berr, Brim, Bruc, Burr, Byng, Cape, Clar, Clev, Alban, Alpha, Bargo, Berry, Bowen, Bulli, Aldgat, Alding, Anakie, Aramac, August, Bamaga, Barham, Barmah, Batlow, Beltan, Bethan, Beulah, Bindoo, Boonah, Bowral, Bright, Buchan, Burnie, Cairns, Casino, Cattai, Adelong, Angasto, Antwerp, Arltung, Ashford, Babinda, Bargara, Barooga, Barraba, Belmont, Bemboka, Benalla, Bendigo, Berrima, Berwick, Beverle, Bicheno, Biloela, Birchip, Bombala, Bonalbo, Brookto, Bruthen, Buderim, Candelo, Capella, Carcoar, Carnama, Chewton, Anglesea, Ardrossa, Armidale, Atherton, Balaklav, Ballarat, Barellan, Belgrave, Berrigan, Binalong, Binnaway, Blackall, Boggabri, Bridport, Brooklyn, Buckland, Bundeena, Bungonia, Busselto, Calliope, Cardwell, Cassilis, Cessnock, Charlton.
Code
plot_forest <- function(data, intercept = TRUE, MA_mean = TRUE){
if (MA_mean == FALSE){
data <- filter(data, Parameter != "overall")
}
p <- ggplot(data, aes(y = estimate,
x = term,
ymin = conf.low,
ymax = conf.high,
shape = parameter_type,
colour = parameter_type)) +
geom_pointrange(fatten = 2) +
ggforestplot::theme_forest() +
theme(axis.line = element_line(linewidth = 0.10, colour = "black"),
axis.line.y = element_blank(),
text = element_text(family = "Helvetica")#,
# axis.text.y = element_blank()
) +
guides(shape = guide_legend(title = NULL),
colour = guide_legend(title = NULL)) +
coord_flip() +
ylab(bquote(Standardised~Effect~Size~Z[r])) +
xlab(element_blank()) +
# scale_y_continuous(breaks = c(-4,-3,-2,-1,0,1),
# minor_breaks = seq(from = -4.5, to = 1.5, by = 0.5)) +
NatParksPalettes::scale_color_natparks_d("Glacier")
if(intercept == TRUE){
p <- p + geom_hline(yintercept = 0)
}
if(MA_mean == TRUE){
p <- p + geom_hline(aes(yintercept = meta_analytic_mean),
data = data,
colour = "#01353D",
linetype = "dashed")
}
return(p)
}
filter_experts <-
rlang::exprs(
exclusion_set == "complete",
estimate_type == "Zr",
model_name == "MA_mod",
publishable_subset == "All",
expertise_subset == "expert")
bt_experts_only <-
ManyEcoEvo_viz %>%
filter(!!!filter_experts,
dataset == "blue tit") %>%
select(tidy_mod_summary) %>%
mutate(plot_data = map(tidy_mod_summary,
.f = ~ dplyr::mutate(.x,
point_shape =
ifelse(stringr::str_detect(term, "overall"),
"diamond",
"circle"),
Parameter =
forcats::fct_reorder(term,
estimate) %>%
forcats::fct_reorder(.,
point_shape,
.desc = TRUE))
),
meta_analytic_mean = map_dbl(plot_data,
~ filter(.x, Parameter == "overall") %>%
pull(estimate))) %>%
select(plot_data, meta_analytic_mean) %>%
unnest(cols = c("plot_data")) %>%
mutate(parameter_type = case_when(str_detect(Parameter, "overall") ~ "mean",
TRUE ~ "study"))
# bt_experts_only <-
# bt_experts_only %>%
# rename(id_col = term) %>%
# group_by(type) %>%
# group_split() %>%
# set_names(., bt_experts_only$type %>% unique) %>%
# # map_if(.x = ., names(.) == "study",
# # .f = ~ anonymise_teams(.x, TeamIdentifier_lookup)) %>%
# bind_rows() %>%
# rename(term = id_col)
bt_forest_experts <- bt_experts_only %>%
arrange(desc(type)) %>%
mutate(type = forcats::as_factor(type)) %>%
group_by(type) %>%
arrange(desc(estimate),.by_group = TRUE) %>%
mutate(term = forcats::as_factor(term),
point_shape = case_when(str_detect(type, "summary") ~ "mean",
TRUE ~ "study")) %>%
plot_forest(intercept = TRUE, MA_mean = TRUE) +
theme(axis.text.x = element_text(size = 15),
axis.title.x = element_text(size = 15),
axis.text.y = element_blank()
) +
scale_y_continuous(limits = c(-1.6, 0.65))
euc_experts_only <-
ManyEcoEvo_viz %>%
filter(!!!filter_experts,
dataset == "eucalyptus") %>%
select(model) %>%
mutate(plot_data = map(model,
.f = ~ broom::tidy(.x,
conf.int = TRUE,
include_studies = TRUE) %>%
dplyr::mutate(point_shape =
ifelse(stringr::str_detect(term, "overall"),
"diamond",
"circle"),
Parameter =
forcats::fct_reorder(term,
estimate) %>%
forcats::fct_reorder(.,
point_shape,
.desc = TRUE))
),
meta_analytic_mean = map_dbl(plot_data,
~ filter(.x, Parameter == "overall") %>%
pull(estimate))) %>%
select(plot_data, meta_analytic_mean) %>%
unnest(cols = c("plot_data")) %>%
mutate(parameter_type = case_when(str_detect(Parameter, "overall") ~ "mean",
TRUE ~ "study"))
# euc_experts_only <-
# euc_experts_only %>%
# rename(id_col = term) %>%
# group_by(type) %>%
# group_split() %>%
# set_names(., euc_experts_only$type %>% unique) %>%
# # map_if(.x = ., names(.) == "study",
# # .f = ~ anonymise_teams(.x, TeamIdentifier_lookup)) %>%
# bind_rows() %>%
# rename(term = id_col)
euc_forest_experts <- euc_experts_only %>%
arrange(desc(type)) %>%
mutate(type = forcats::as_factor(type)) %>%
group_by(type) %>%
arrange(desc(estimate),.by_group = TRUE) %>%
mutate(term = forcats::as_factor(term),
point_shape = case_when(str_detect(type, "summary") ~ "mean",
TRUE ~ "study")) %>%
plot_forest(intercept = TRUE, MA_mean = TRUE) +
theme(axis.text.x = element_text(size = 15),
axis.title.x = element_text(size = 15),
axis.text.y = element_blank()
) +
scale_y_continuous(limits = c(-5, 1),
breaks = c(-5, -4, -3, -2, -1, 0, 1) )
# ---- Extract Viz ----
bt_forest_experts
euc_forest_experts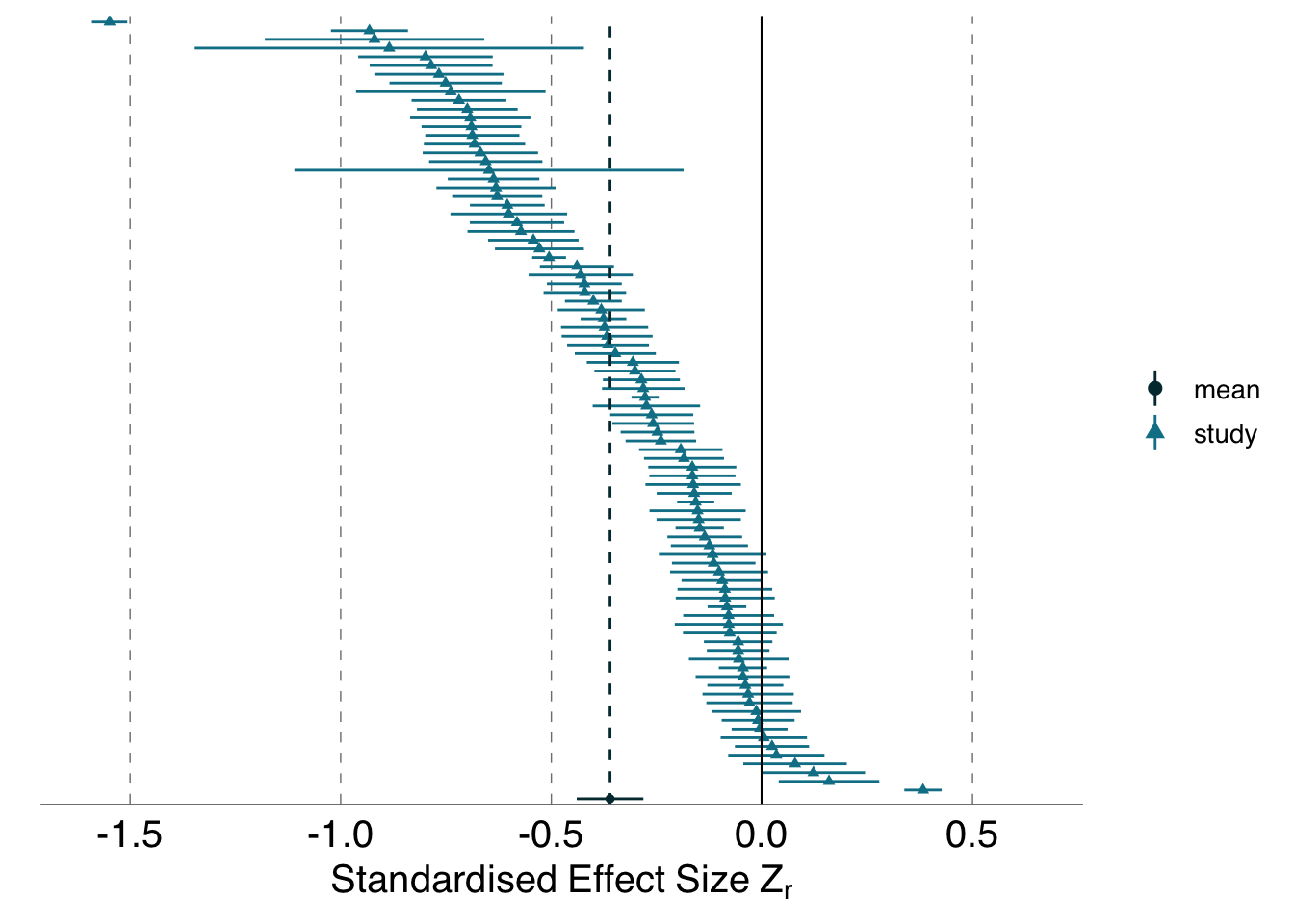

B.1.1.5 Post-hoc analysis: Exploring the effect of excluding analyses of the blue tit dataset containing highly collinear predictor variables
For the blue tit dataset, we created a subset of analyses that excluded effects based on analyses containing highly correlated predictor variables. Excluded analyses are exported internally in the ManyEcoEvo package as ManyEcoEvo::collinearity_subset. Analyses with the following identifiers are excluded in the reduced subset: Armadal-1-1-1, Babinda-1-1-1, Babinda-2-2-1, Barham-1-1-1, Barham-2-2-1, Bega-1-1-1, Bega-1-1-2, Bega-2-2-1, Bega-2-2-2, Borde-1-1-1, Bruc-1-1-1, Caigun-1-1-1, Caigun-2-2-1, Adelong-1-1-1, Adelong-2-2-1.
Code
filter_collinear <- rlang::exprs(exclusion_set == "complete",
publishable_subset == "All",
expertise_subset == "All",
collinearity_subset == "collinearity_removed",
model_name == "MA_mod",
dataset == "blue tit")
# summary_output_params <- rlang::exprs(tidy_mod_summary, MA_fit_stats, mod_fit_stats)
ManyEcoEvo_viz %>%
filter(!!!filter_collinear) %>%
mutate(plot_data = map(tidy_mod_summary,
.f = ~ dplyr::mutate(.x,
point_shape =
ifelse(stringr::str_detect(term, "overall"),
"diamond",
"circle"),
Parameter =
forcats::fct_reorder(term,
estimate) %>%
forcats::fct_reorder(.,
point_shape,
.desc = TRUE))
),
meta_analytic_mean = map_dbl(plot_data,
~ filter(.x, Parameter == "overall") %>%
pull(estimate))) %>%
select(plot_data, meta_analytic_mean) %>%
unnest(cols = c("plot_data")) %>%
mutate(parameter_type = case_when(str_detect(Parameter, "overall") ~ "mean",
TRUE ~ "study")) %>%
arrange(desc(type)) %>%
mutate(type = forcats::as_factor(type)) %>%
group_by(type) %>%
arrange(desc(estimate),.by_group = TRUE) %>%
mutate(term = forcats::as_factor(term),
point_shape = case_when(str_detect(type, "summary") ~ "mean",
TRUE ~ "study")) %>%
plot_forest(intercept = TRUE, MA_mean = TRUE) +
theme(axis.text.x = element_text(size = 15),
axis.title.x = element_text(size = 15),
axis.text.y = element_blank()
) +
scale_y_continuous(limits = c(-1.5, 0.5),
breaks = c(-1.5, -1, -0.5, 0, 0.5) )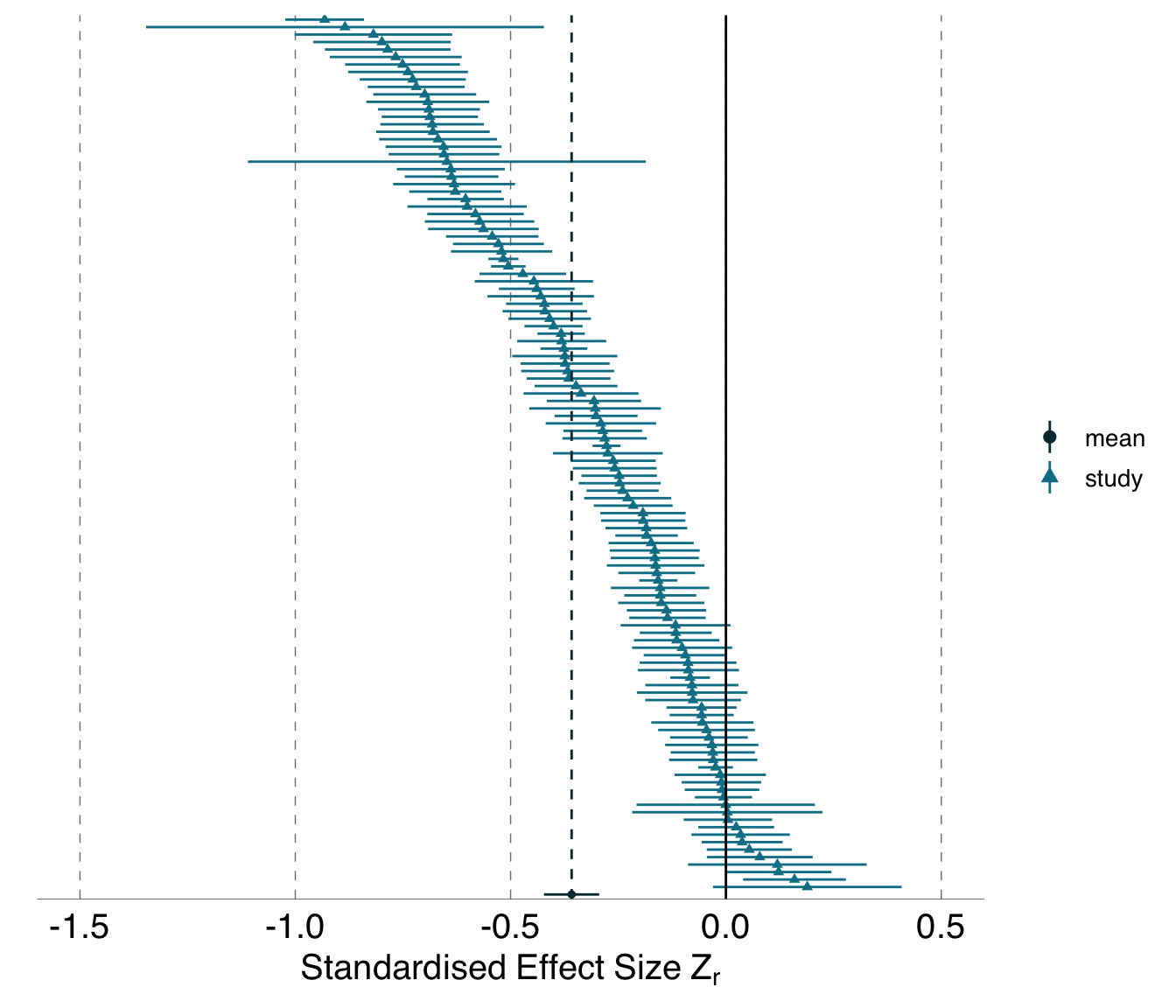
B.1.2 Out of sample predictions \(y_i\)
B.1.2.1 Excluded analyses with constructed variables
Code
by <- join_by(response_variable_name) # don't join on id_col: inc. other excl.
# Analyst Constructed Variables
all_constructed_vars <-
ManyEcoEvo %>%
pull(data, dataset) %>%
list_rbind(names_to = "dataset") %>%
filter(str_detect(response_variable_type, "constructed")) %>%
distinct(dataset,response_variable_name) %>%
drop_na() %>%
arrange()
# Constructed Variables Included in the ManyAnalysts meta-analysis
# (i.e. we have included them in the parameter tables)
ManyEcoEvo_yi_constructed_vars <-
ManyEcoEvo:::analysis_data_param_tables %>%
distinct(variable, dataset) %>%
rename(response_variable_name = variable) %>%
semi_join(all_constructed_vars, by) %>%
filter(!str_detect(response_variable_name,
"average.proportion.of")) # was excluded
yi_constructed <-
ManyEcoEvo_yi_results %>%
pull(data, dataset) %>%
list_rbind(names_to = "dataset") %>%
filter(str_detect(response_variable_type, "constructed")) %>%
distinct(dataset, id_col, TeamIdentifier, response_variable_name) %>%
drop_na()
excluded_yi_constructed <-
ManyEcoEvo %>%
pull(data, dataset) %>%
list_rbind(names_to = "dataset") %>%
filter(str_detect(response_variable_type, "constructed"),
str_detect(exclusions_all, "retain")) %>%
distinct(dataset, id_col, TeamIdentifier, response_variable_name) %>%
drop_na() %>%
anti_join(yi_constructed, by) #rm response vars in yi_constructed
n_dropped_analyses <-
excluded_yi_constructed %>%
n_distinct("id_col")
n_teams_w_dropped_analyses <-
excluded_yi_constructed %>%
group_by(TeamIdentifier) %>%
count() %>%
n_distinct("TeamIdentifier")We standardized the \(y_i\) estimates and their standard errors for the blue tit analyses using the population mean and standard deviations of the corresponding dependent variable for that analysis, as shown in Equation B.1, using the function ManyEcoEvo::Z_VZ_preds(). Note that this is NOT the same process as standardizing the effect sizes \(Z_r\). We used the mean and standard deviation of the relevant raw datasets as our ‘population’ parameters.
\[ Z_j = \frac{\mu_i-\bar{x}_j}{\text{SD}_j} \\ \\ {\text{VAR}}_{Z_j} = \frac{{\text{SE}}_{\mu_i}}{{\text{SD}_j}} \\ \tag{B.1}\]
Where \(\mu\) is the population parameter taken from our original dataset for variable \(i\), and \(\bar{x}_j\) and \(\text{SD}_j\) are the out of sample point estimate values supplied for analysis \(j\). \(\text{SE}_{{\mu}_{i}}\) is the standard error of the population mean for variable \(i\), while \({\text{VAR}}_{{Z}_{j}}\) and \({Z}_{j}\) are the standardized variance and mean estimate for analysis \(j\). Note that for the response variables that were scaled-and-centered, or else mean-centred before model fitting, we do not need to standardise because these are already on the Z-scale. In doing so we make the assumption that analysts’ data subsetting will have little effect on the outcomes. For some analyses of the blue tit dataset, analysts constructed their own unique response variables, which meant we needed to reconstruct these variables in order to calculate the population parameters. Unfortunately we were not able to re-construct all variables used by the analysts, as we were unable to reproduce the data required for their re-construction, e.g. we were unable to reproduce principal component analyses or fitted models for extracting residuals Table B.2. A total of 15 were excluded from out-of-sample meta-analysis, from 10 teams, including the following analysis identifiers: Bruc-1-1-1, Clar-2-2-1, Clar-1-1-1, Batlow-1-1-1, Batlow-1-1-2, Bindoo-1-1-1, Bourke-1-1-1, Buchan-1-1-1, Arltung-4-4-1, Bargara-1-1-1, Bendigo-5-5-1, Booligal-3-3-1, Booligal-2-2-1, Booligal-4-4-1 and Booligal-5-5-1.
Code
all_constructed_vars %>%
semi_join(ManyEcoEvo_yi_constructed_vars, by) %>%
mutate(included_in_yi = TRUE) %>%
bind_rows(
{
all_constructed_vars %>%
anti_join(ManyEcoEvo_yi_constructed_vars, by) %>%
mutate(included_in_yi = FALSE)
}
) %>%
dplyr::mutate(included_in_yi =
case_match(included_in_yi,
TRUE ~ "check",
FALSE ~ "xmark" ),
response_variable_name =
gluedown::md_code(response_variable_name)) %>%
group_by(dataset) %>%
gt::gt() %>%
gt::cols_label(response_variable_name = "Constructed Variable",
included_in_yi = gt::md("Variable reconstructed for meta-analysis?")) %>%
gt::fmt_icon(included_in_yi) %>%
gt::tab_style(style = cell_text(style = "italic", transform = "capitalize"),
locations = cells_row_groups(groups = "eucalyptus")) %>%
gt::tab_style(style = cell_text(align = "center"),
locations = cells_body(columns = included_in_yi)) %>%
gt::tab_style(style = cell_text(align = "left"),
locations = cells_body(columns = response_variable_name)) %>%
gt::tab_style(style = cell_text(align = "left"),
locations = cells_column_labels(response_variable_name)) %>%
gt::tab_style(locations = cells_body(columns = response_variable_name),
style = cell_text(size = "small")) %>%
gt::fmt_markdown(columns = response_variable_name) %>%
gt::opt_stylize(style = 6, color = "gray", add_row_striping = TRUE) %>%
gt::opt_row_striping(row_striping = TRUE) | Constructed Variable | Variable reconstructed for meta-analysis? |
|---|---|
| blue tit | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| eucalyptus | |
|
|
|
|
|
|
|
|
|
|
B.1.2.2 Non-truncated \(y_{i}\) meta-analysis forest plot
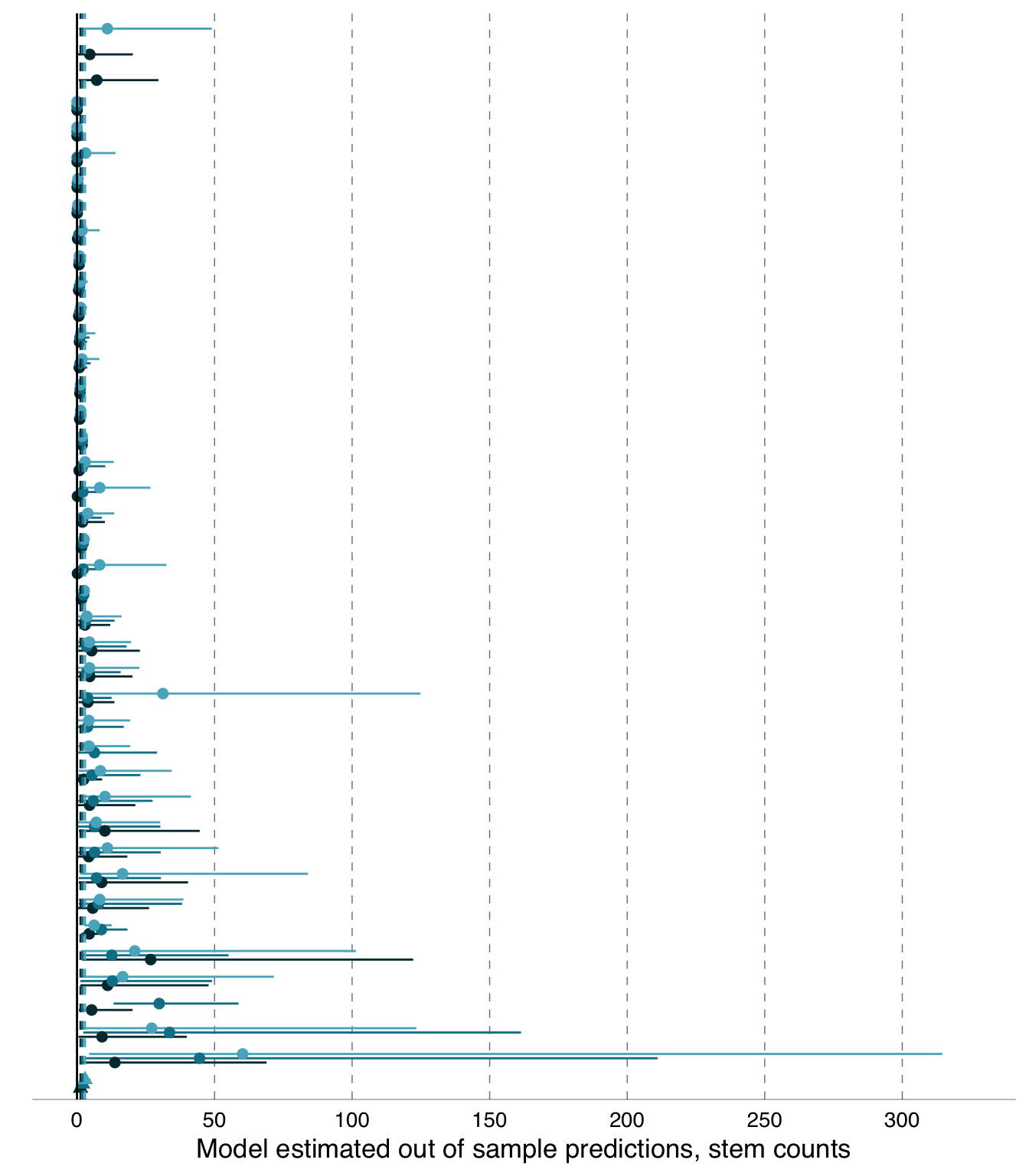
Below is the non-truncated version of Figure 3.3 showing a forest plot of the out-of-sample predictions, \(y_{i}\), on the response-scale (stem counts), for Eucalyptus analyses, showing the full error bars of all model estimates.
Code
plot_forest_2 <- function(data, intercept = TRUE, MA_mean = TRUE, y_zoom = numeric(2L)){
if(MA_mean == FALSE){
data <- filter(data, study_id != "overall")
}
plot_data <- data %>%
group_by(study_id) %>%
group_nest() %>%
hoist(data, "estimate",.remove = FALSE) %>%
hoist(estimate, y50 = 2) %>%
select(-estimate) %>%
unnest(data) %>%
arrange(desc(type)) %>%
mutate(type = forcats::as_factor(type)) %>%
group_by(type) %>%
arrange(desc(y50),.by_group = TRUE) %>%
mutate(study_id = forcats::as_factor(study_id),
point_shape = case_when(str_detect(type, "summary") ~ "diamond",
TRUE ~ "circle"))
p <- ggplot(plot_data, aes(y = estimate,
x = study_id,
ymin = conf.low,
ymax = conf.high,
# shape = type,
shape = point_shape,
colour = estimate_type
)) +
geom_pointrange(position = position_dodge(width = 0.5)) +
ggforestplot::theme_forest() +
theme(axis.line = element_line(linewidth = 0.10, colour = "black"),
axis.line.y = element_blank(),
text = element_text(family = "Helvetica")) +
guides(shape = "none", colour = "none") +
coord_flip(ylim = y_zoom) +
labs(y = "Model estimated out of sample predictions, stem counts",
x = element_blank()) +
scale_y_continuous(breaks = scales::breaks_extended(10)) +
NatParksPalettes::scale_color_natparks_d("Glacier")
if(intercept == TRUE){
p <- p + geom_hline(yintercept = 0)
}
if(MA_mean == TRUE){
p <- p +
geom_hline(aes(yintercept = plot_data %>%
filter(type == "summary", estimate_type == "y25") %>%
pluck("estimate")),
data = data,
colour = "#01353D",
linetype = "dashed") +
geom_hline(aes(yintercept = plot_data %>%
filter(type == "summary", estimate_type == "y50") %>%
pluck("estimate")),
data = data,
colour = "#088096",
linetype = "dashed") +
geom_hline(aes(yintercept = plot_data %>%
filter(type == "summary", estimate_type == "y75") %>%
pluck("estimate")),
data = data,
colour = "#58B3C7" ,
linetype = "dashed")
}
print(p)
}
# ---- new code ----
eucalyptus_yi_plot_data <-
ManyEcoEvo_yi_viz %>%
filter(dataset == "eucalyptus", model_name == "MA_mod") %>%
unnest(cols = tidy_mod_summary) %>%
mutate(response_scale = list(log_back(estimate, std.error, 1000)),
.by = c(dataset, estimate_type, term, type),
.keep = "used") %>%
select(-estimate, -std.error) %>%
unnest_wider(response_scale) %>%
rename(estimate = mean_origin, conf.low = lower, conf.high = upper) %>%
nest(tidy_mod_summary = c(-dataset, -estimate_type)) %>% #extract euc data for plotting (on count scale, not log scale)
select(dataset, estimate_type, tidy_mod_summary) %>%
unnest(cols = tidy_mod_summary) %>%
rename(study_id = term) %>%
ungroup()
max_x_axis <-
eucalyptus_yi_plot_data %>%
pluck("conf.high", max) %>%
round() + 10
eucalyptus_yi_plot_data %>%
plot_forest_2(MA_mean = T, y_zoom = c(0, max_x_axis)) +
theme(axis.text.y = element_blank())